Clustering with FSharp.Stats II: hierarchical clustering¶
Summary: This tutorial demonstrates hierarchical clustering with FSharp.Stats and how to visualize the results with Plotly.NET.
In the previous article of this series k-means clustering using FSharp.Stats was introduced.
Introduction¶
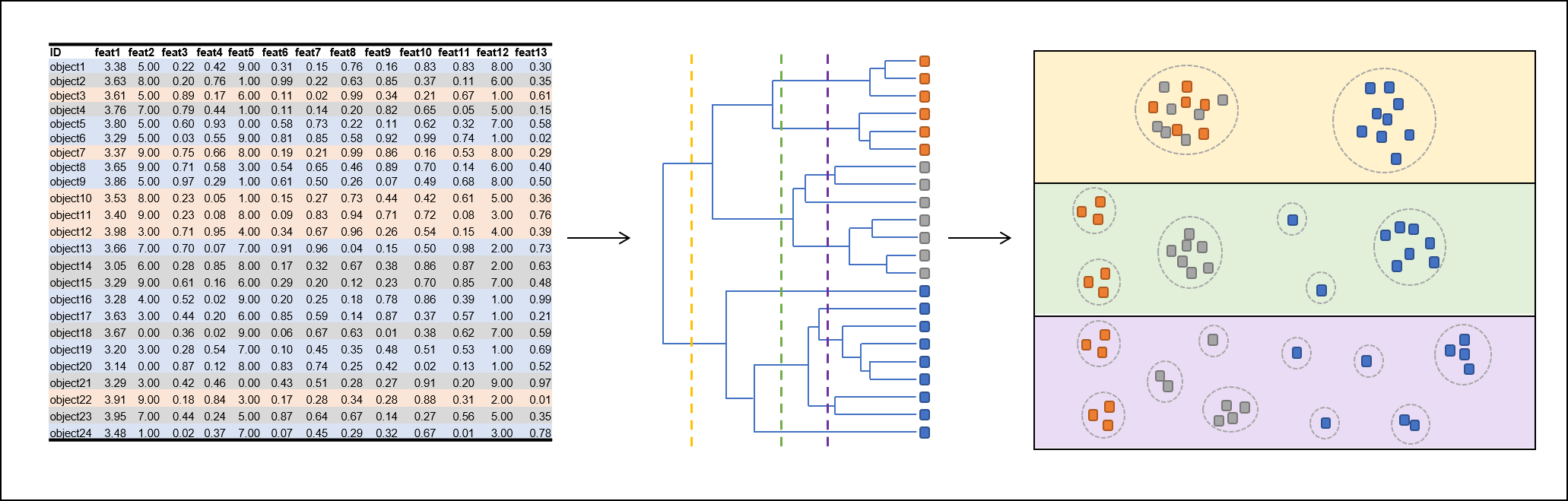
Clustering methods can be used to group elements of a huge data set based on their similarity. Elements sharing similar properties cluster together and can be reported as coherent group. Many clustering algorithms require a predefined cluster number, that has to be provided by the experimenter. Hierarchical clustering (hClust) does not require such cluster number definition. Instead, hierarchical clustering results in a tree structure, that has a single cluster (node) on its root and recursively splits up into clusters of elements that are more similar to each other than to elements of other clusters. For generating multiple clustering results with different number of clusters, the clustering has to performed only once. Subsequently a cluster number can be defined to split up the clustering tree in the desired number of clusters. The clustering tree is often represented as dendrogram.
There are two types of hClust:¶
Agglomerative (bottom-up): Each data point is in its own cluster and the nearest ones are merged recursively. It is referred as agglomerative hierarchical clustering.
Divisive (top-down): All data points are in the same cluster and you divide the cluster into two that are far away from each other.
The presented implementation is an agglomerative type.
Distance measures¶
There are several distance metrics, that can be used as distance function. The commonly used one probably is Euclidean distance.
Linker¶
When the distance between two clusters is calculated, there are several linkage types to choose from:
complete linkage: maximal pairwise distance between the clusters (prone to break large clusters)
single linkage: minimal pairwise distance between the clusters (sensitive to outliers)
centroid linkage: distance between the two cluster centroids
average linkage: average pairwise distance between the clusters (sensitive to cluster shape and size)
median linkage: median pairwise distance between the clusters
For demonstration of hierarchical clustering, the classic iris data set is used, which consists of 150 records, each of which contains four measurements and a species identifier.
Referencing packages¶
#r "nuget: Deedle.Interactive, 3.0.0"
#r "nuget: FSharp.Stats, 0.4.3"
#r "nuget: Plotly.NET.Interactive, 4.0.0"
#r "nuget: FSharp.Data, 4.2.7"
- Deedle.Interactive, 3.0.0
- FSharp.Data, 4.2.7
- FSharp.Stats, 0.4.3
- Plotly.NET.Interactive, 4.0.0
Loading extensions from `C:\Users\schne\.nuget\packages\plotly.net.interactive\4.0.0\interactive-extensions\dotnet\Plotly.NET.Interactive.dll`
Loading extensions from `C:\Users\schne\.nuget\packages\deedle.interactive\3.0.0\interactive-extensions\dotnet\Deedle.Interactive.dll`
Loading data¶
open FSharp.Data
open Deedle
// Retrieve data using the FSharp.Data package and read it as dataframe using the Deedle package
let rawData = Http.RequestString @"https://raw.githubusercontent.com/fslaborg/datasets/main/data/iris.csv"
let df = Frame.ReadCsvString(rawData)
df
| sepal_length | sepal_width | petal_length | petal_width | species | (Decimal) | (Decimal) | (Decimal) | (Decimal) | (string) |
|---|---|---|---|---|---|---|
| 0 | -> | 5.5 | 2.4 | 3.8 | 1.1 | versicolor |
| 1 | -> | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
| 2 | -> | 7.6 | 3 | 6.6 | 2.1 | virginica |
| 3 | -> | 5.6 | 2.8 | 4.9 | 2 | virginica |
| 4 | -> | 6.1 | 3 | 4.9 | 1.8 | virginica |
| : | ... | ... | ... | ... | ... | |
| 145 | -> | 7.7 | 2.6 | 6.9 | 2.3 | virginica |
| 146 | -> | 5.7 | 2.6 | 3.5 | 1 | versicolor |
| 147 | -> | 5.9 | 3 | 5.1 | 1.8 | virginica |
| 148 | -> | 6.8 | 3.2 | 5.9 | 2.3 | virginica |
| 149 | -> | 5 | 3.6 | 1.4 | 0.2 | setosa |
150 rows x 5 columns
0 missing values
Let's take a first look at the data with heatmaps using Plotly.NET. Each of the 150 records consists of four measurements and a species identifier. Since the species identifier occur several times (Iris-virginica, Iris-versicolor, and Iris-setosa), we create unique labels by adding the rows index to the species identifier.
open Plotly.NET
let colNames = ["sepal_length";"sepal_width";"petal_length";"petal_width"]
// isolate data as float [] []
let data =
Frame.dropCol "species" df
|> Frame.toJaggedArray
// isolate labels as seq<string>
let labels =
Frame.getCol "species" df
|> Series.values
|> Seq.mapi (fun i s -> sprintf "%s_%i" s i)
|> Array.ofSeq
Chart.Heatmap(data,colNames=colNames,rowNames=labels)
// required to fit the species identifier on the left side of the heatmap
|> Chart.withMarginSize(Left=100.)
|> Chart.withTitle "raw iris data"
Clustering¶
The function that performs hierarchical clustering can be found at FSharp.Stats.ML.Unsupervised.HierarchicalClustering.generate. It requires three input parameters:
- Distance measure working on
'T(fromFSharp.Stats.ML.DistanceMetrics) - Linkage type
- Data to cluster as
'T
open FSharp.Stats.ML
open FSharp.Stats.ML.Unsupervised
let distanceMeasure = DistanceMetrics.euclideanNaNSquared
let linker = HierarchicalClustering.Linker.centroidLwLinker
// calculates the clustering and reports a single root cluster (node),
// that may recursively contains further nodes
let clusterResultH =
HierarchicalClustering.generate distanceMeasure linker data
// If a desired cluster number is specified, the following function cuts the cluster according
// to the depth, that results in the respective number of clusters (here 3). Only leaves are reported.
let threeClusters = HierarchicalClustering.cutHClust 3 clusterResultH
Every cluster leaf contains its raw values and an index that indicates the position of the respective data point in the raw data. The index can be retrieved from leaves using HierarchicalClustering.getClusterId.
// Detailed information for 3 clusters are given
let inspectThreeClusters =
threeClusters
|> List.map (fun cluster ->
cluster
|> List.map (fun leaf ->
labels.[HierarchicalClustering.getClusterId leaf]
)
)
inspectThreeClusters
|> List.mapi (fun i x ->
let truncCluster = x.[0..4] |> String.concat "; "
sprintf "Cluster%i: [%s ...]" i truncCluster
)
|> String.concat "\n"
Cluster0: [versicolor_44; versicolor_76; versicolor_89; versicolor_12; versicolor_93 ...] Cluster1: [setosa_16; setosa_36; setosa_74; setosa_42; setosa_122 ...] Cluster2: [virginica_72; virginica_135; virginica_14; virginica_7; virginica_73 ...]
To break up the tree structure but maintain the clustering order, the cluster tree has to be flattened.
// To recursevely flatten the cluster tree into leaves only, use flattenHClust.
// A leaf list is reported, that does not contain any cluster membership,
// but is sorted by the clustering result.
let hLeaves =
clusterResultH
|> HierarchicalClustering.flattenHClust
// Takes the sorted cluster result and reports a tuple of label and data value.
let dataSortedByClustering =
hLeaves
|> Seq.choose (fun c ->
let label = labels.[HierarchicalClustering.getClusterId c]
let values = HierarchicalClustering.tryGetLeafValue c
match values with
| None -> None
| Some x -> Some (label,x)
)
The visualization again is performed using a Plotly.NET heatmap.
open FSharpAux
let (hlable,hdata) =
dataSortedByClustering
|> Seq.unzip
Chart.Heatmap(hdata,colNames=colNames,rowNames=hlable)
// required to fit the species identifier on the left side of the heatmap
|> Chart.withMarginSize(Left=100.)
|> Chart.withTitle "Clustered iris data (hierarchical clustering)"
Limitations¶
- There is no strong guidance on which distance function and linkage type should be used. It often is chosen arbitrarily according to the user's experience.
- The visual interpretation of the dendrogram is difficult, since swapping the direction of some bifurcations may totally disturbe the visual impression.
Notes¶
- Please note that depending on what data you want to cluster, a column wise z-score normalization may be required. In the presented example differences in sepal width have a reduced influence because the absolute variation is low.
References¶
- Vijaya et al., A Review on Hierarchical Clustering Algorithms, Journal of Engineering and Applied Sciences, 2017
- Rani and Rohil, A Study of Hierarchical Clustering Algorithm, International Journal of Information and Computation Technology, 2013
- FSharp.Stats documentation, fslaborg, https://fslab.org/FSharp.Stats/Clustering.html
Further reading¶
Examples are taken from FSharp.Stats documentation that covers various techniques for an optimal cluster number determination.
The next article in this series covers DBSCAN using FSharp.Stats.